Data Description¶
The LNDb dataset contains 294 CT scans collected retrospectively at the Centro Hospitalar e Universitário de São João (CHUSJ) in Porto, Portugal between 2016 and 2018. All data was acquired under approval from the CHUSJ Ethical Commitee and was anonymised prior to any analysis to remove personal information except for patient birth year and gender. Further details on patient selection and data acquisition can be consulted on the database description paper.
Each CT scan was read by at least one radiologist at CHUSJ to identify pulmonary nodules and other suspicious lesions. A total of 5 radiologists with at least 4 years of experience reading up to 30 CTs per week participated in the annotation process throughout the project. Annotations were performed in a single blinded fashion, i.e. a radiologist would read the scan once and no consensus or review between the radiologists was performed. Each scan was read by at least one radiologist. The instructions for manual annotation were adapted from LIDC-IDRI. Each radiologist identified the following lesions:
- nodule ⩾3mm: any lesion considered to be a nodule by the radiologist with greatest in-plane dimension larger or equal to 3mm;
- nodule <3mm: any lesion considered to be a nodule by the radiologist with greatest in-plane dimension smaller than 3mm;
- non-nodule: any pulmonary lesion considered not to be a nodule by the radiologist, but that contains features which could make it identifiable as a nodule;
The annotation process varied for the different categories. Nodules ⩾3mm were segmented and subjectively characterized according to LIDC-IDRI (ratings on subtlety, internal structure, calcification, sphericity, margin, lobulation, spiculation, texture and likelihood of malignancy). For a complete description of these characteristics the reader is referred to McNitt-Gray et al.. For nodules \<3mm the nodule centroid was marked and subjective assessment of the nodule's characteristics was performed. For non-nodules, only the lesion centroid was marked. Given that different radiologists may have read the same CT and no consensus review was performed, variability in radiologist annotations is expected.
Note that from the 294 CTs of the LNDb dataset, 58 CTs with annotations by at least two radiologists have been withheld for the test set, as well as the corresponding annotations.Instructions on how to download the LNDb dataset can be found at the Download page.
Data Formats¶
CT data is available on MetaImage (.mhd/.raw) format. A script for reading .mhd/.raw files is available for download (utils.py). Filenames follow the format LNDb-XXXX.mhd where XXXX is the LNDb CT ID.
Individual nodule annotations are available on a csv file (trainNodules.csv) that contains one finding marked by a radiologist per line. Each line holds the LNDb CT ID, the radiologist that marked the finding (numbered from 1 to nrad within each CT), the finding's ID (numbered from 1 to nfinding within each CT for each radiologist), the xyz coordinates of the finding in world coordinates, whether it is a nodule (1) or a non-nodule (0), the corresponding nodule volume and the nodule texture rating given (1-5). For non-nodules, the texture given is 0.
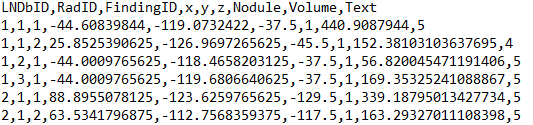
The list of nodule annotations after merging the annotations of different radiologists is available on separate a csv file (trainNodules_gt.csv) that contains one finding per line. Each line holds the LNDb CT ID, the radiologists that marked the finding (numbered from 1 to nrad within each CT), the ID of the matching finding for each radiologist on trainNodules.csv, the unique nodule ID after merging (numbered from 1 to nfinding within each CT), the xyz coordinates of the finding in world coordinates, the agreement level (number of radiologists that annotated each finding, whether it is a nodule (1) or a non-nodule (0), the corresponding nodule volume and the nodule texture (average of texture ratings given). For non-nodules, the texture given is 0.

Nodule segmentations are given on MetaImage (*.mhd/*.raw) format. Each LNDbXXXX_radR.mhd holds the segmentation for all nodules on CT XXXX according to radiologist R in a 3D array of the CT's size where the value of each pixel is the finding's ID in trainNodules.csv.
Finally, Fleischner scores are available on a separate csv file (trainFleischner.csv) that contains one scan per line. Each line holds the LNDb CT ID and the ground truth Fleischner score.

External Data¶
The use of data other than the LNDb dataset, public or otherwise, is fully allowed. However, please disclose any data used when submitting your ICIAR 2020 conference paper.
